Services

DNA/RNA isolation
Hartwig Sequencing Services is specialized in DNA and RNA isolation for all types of fresh frozen tissues, blood, and cell pellets. We use the QIAsymphony system to perform DNA and RNA isolation. Please contact us for more information.
DNA/RNA library preparation
DNA
Amplified and PCR-free
Samples requiring DNA library preparation are processed on our unique in house programmed robotic set up, based on the Biomek i7 liquid handler system. This setup is scalable from one to 88 samples per day. We use the Enzymatic WGS Library prep KAPA EvoPlus kit for our library prep, which is based on ligation and fragmentation by Enzymatic reactions. Our library preparation method with amplification is validated and optimized for gDNA standard input of 50 – 70 ng.
RNA
Ribo depletion
Samples requiring RNA library preparation are processed on our unique in house programmed robotic set up, based on the Biomek 4000 liquid handler system. This setup is scalable from one to 24 samples per day. We use the KAPA RNA HyperPrep Kit with RiboErase (HMR). Our RNA library preparation method is validated and optimized for a standard input of 100 and 200 ng total RNA in 10 µl.
See the Sample submission guide for our Sequencing Services. Please contact us if your request has other sample specifications than described in the Sample submission guide.


Next Generation Sequencing
Hartwig Sequencing Services is available for libraries prepared by us or ready-to-sequence library pools from customers. Sequencing can also be used for QC only.
We have the following Illumina-based DNA sequencing systems available:
- NovaSeq6000
- iSeq100
SNP Genotyping
Hartwig Sequencing Services uses SNP Genotyping for human sample identification and tracking. The QuantStudio 12K Flex, a TaqMan based fingerprinting platform, is available and results can be used for comparison with human WGS sequencing data. This is an effective method to validate and identify samples, for example in biobanks or tissue culture.
"The rapid turnover of our own sequencing as well as the clear access policy to the Hartwig Medical database enabled rapid progression of our project and successful publication without delay."
Hans Clevers, group leader — Hubrecht Institute

Bioinformatics
Hartwig Sequencing Services provides limited bioinformatics analysis and support. Almost all of our customers receive FASTQ data and subsequently do their own analysis.
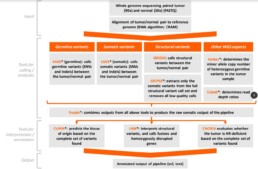
High-end WGS based somatic DNA analysis
Some of our customers use our in-house developed high-end WGS based somatic DNA analysis pipeline instead. The somatic pipeline is under continuously development to include the latest insights that genomics offers oncology. We offer a comprehensive somatic DNA analysis pipeline, mapping of sequencing data.
Unaligned sequence read data-files
We can also provide you with a collection of unaligned sequence read data-files. All of our software is open source and can be deployed on any sufficiently powerful computer. We provide these analyses as a service as well.
The somatic DNA analysis pipeline is for data derived from fresh frozen samples only and optimized for 90x tumor vs 30x normal. Availability of a paired normal sample is mandatory. Main features include:
- tumor purity estimation
- SNV, MNV, CNV, INDELS, SV, TML and MSI
- sensitive, precise and consistent tumor-adjusted copy number and structural variant determination
- new insights for short structural variants and regions of complex local topology
Click to enlarge the somatic DNA analysis pipeline flow chart.
Please swipe left and right to see full table
| Input material | Amount | Quality | Application |
|---|---|---|---|
| Fresh Frozen tissue*** | >1mm2 | DNA/RNA isolation | |
| Cell pellets** | >100.000 cells | DNA/RNA isolation | |
| gDNA (amplification)* | >500ng (10ng/µl) | OD 260/280 = 1.8-2.0 | Library preparation |
| RNA (ribo)** | >200ng (10ng/µl) | RIN >6 | Library Preparation |
| NGS library pool* | >400µl | >5nM | NovaSeq S4 |
| NGS library pool* | >200µl | >5nM | NovaSeq S2 |
| NGS library pool* | >100µl | >5nM | NovaSeq S1 |
| NGS library pool* | >25µl | >5nM | NextSeq |
| NGS library pool* | >300µl | >5nM | iSeq – NovaSeq |
| NGS library pool* | >25µl | >5nM | iSeq |
* Dilute samples in FluidX 1mL tubes (P/N:68-1002-10) with 10 mM Tris/1 mM EDTA or similar standard elution buffer and send at 4 °C.
** Send in marked 1.5mL Eppendorf tube on dry ice.
*** Send in scannable barcoded tissue container on dry ice.
Quantify gDNA or RNA samples with dsDNA specific or RNA specific fluorescent method (e.g.
Qubit) and quantify NGS libraries with qPCR or dsDNA specific fluorescent method in combination with BioAnalyzer. Please contact us if your request has other sample specifications than described in the Sample submission guide.